Last updated: 5th Dec, 2023
The class imbalance problem in machine learning occurs when the classes in a dataset are not represented equally, leading to a significant difference in the number of instances for different classes. This imbalance can cause a classification model to be biased towards the majority class, resulting in poor performance on the minority class. Thus, the class imbalance hinders data scientists by challenging the development of accurate and fair models, as the skewed distribution can lead to misleading training predictions / outcomes and reduced effectiveness in real-world applications where minority classes are critical.
In this post, you will learn about how to tackle class imbalance issue when training machine learning classification models with imbalanced dataset. This is illustrated using Python SKlearn example. In the same context, you may check out my earlier post on handling class imbalance using class_weight. As a data scientist, it is of utmost importance to learn some of these techniques as you will often come across the class imbalance problem while working on different classification problems.
Creating Class Imbalanced Dataset using Sklearn Resample
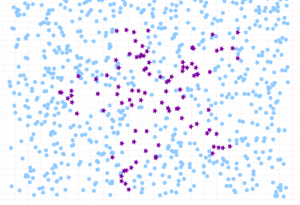
Here is how the class imbalance in the dataset can be visualized:
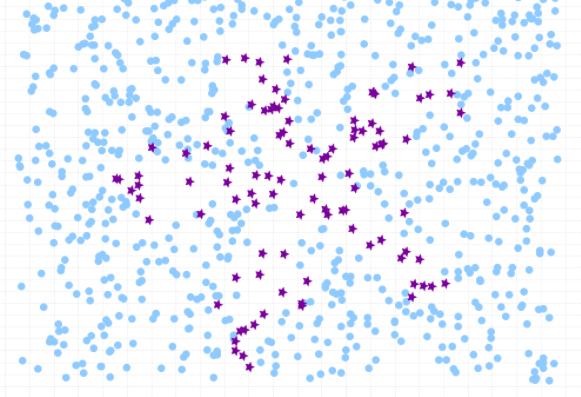
Before going ahead and looking at the Python code example related to how to use Sklearn.utils resample method, lets create an imbalanced data set having class imbalance. We will create imbalanced dataset with Sklearn breast cancer dataset. Here is the Python code sample. In the code below, the resample method from sklearn.utils is used to downsample the malignant class to only 30 instances. Then, the downsampled malignant instances is combined with the benign instances to create an imbalanced dataset.
import numpy as np from sklearn.datasets import load_breast_cancer from sklearn.utils import resample # Load the breast cancer dataset data = load_breast_cancer() X = data.data y = data.target # Separate the dataset into benign and malignant X_benign = X[y == 1] X_malignant = X[y == 0] # Downsample the malignant class to have only 30 instances X_malignant_downsampled = resample(X_malignant, n_samples=30, random_state=42) # Combine the downsampled malignant class with the benign class X_imbalanced = np.vstack((X_benign, X_malignant_downsampled)) y_imbalanced = np.hstack((np.ones(X_benign.shape[0]), np.zeros(X_malignant_downsampled.shape[0]))) # Verify the counts of each class benign_count = np.sum(y_imbalanced == 1) malignant_count = np.sum(y_imbalanced == 0) (benign_count, malignant_count)
The Python code results in creating an imbalanced dataset with 212 records labeled as malignant class reduced to 30. Thus, the total records count becomes benign tumour (357) + malignant tumour (30).
Next step is to use resample method to oversample the minority class (malignant tumour records in this example) and undersample the majority class (benign tumour records).
Resample method for Oversampling or Upsampling Minority Class
The idea is to oversample or upsample the data related to minority class using replacement. This refers to the process of increasing the number of instances in the minority class to balance the class distribution. One of the parameter in resample method is replace and other one is n_samples which relates to number of samples to which minority class will be oversampled. In addition, you can also use stratify to create sample in the stratified fashion. Once the sampling is done, the balanced dataset is created by appending the oversampled dataset. Here is the code representing the following aspects:
- Oversampling of minority class
- Creating balanced data set by appending the oversampled dataset
import numpy as np
from sklearn.utils import resample
#
# Create oversampled training data set for minority class
#
X_oversampled, y_oversampled = resample(X_imbalanced[y_imbalanced == 0],
y_imbalanced[y_imbalanced == 0],
replace=True,
n_samples=X_imbalanced[y_imbalanced == 1].shape[0],
random_state=123)
# Append the oversampled minority class to the imbalanced data and related labels
X_balanced = np.vstack((X_imbalanced[y_imbalanced == 1], X_oversampled))
y_balanced = np.hstack((y_imbalanced[y_imbalanced == 1], y_oversampled))
Once balanced dataset is created using oversampling of minority class, the model training is carried out in the usual manner. Here is the rest of the code for training. Note that the code below used the following steps for training and scoring the model:
- Creating training and test split
- Create the pipeline
- Create a randomized search (RandomizedSearchCV) for model tuning
- Fit the randomizedSearchCV estimator
- Score the model
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
from sklearn.metrics import accuracy_score
from sklearn.model_selection import RandomizedSearchCV
import scipy as sc
#
# Create training and test split using the balanced dataset
# created by oversampling
#
X_train, X_test, y_train, y_test = train_test_split(X_balanced, y_balanced, test_size=0.3,
random_state=1, stratify=y_balanced)
#
# Create the pipeline
#
pipeline = make_pipeline(StandardScaler(), LogisticRegression(random_state=1))
#
# Create the randomized search estimator
#
param_distributions = [{'logisticregression__C': sc.stats.expon(scale=100)}]
rs = RandomizedSearchCV(estimator=pipeline, param_distributions = param_distributions,
cv = 10, scoring = 'accuracy', refit = True, n_jobs = 1,
random_state=1)
#
# Fit the model
#
rs.fit(X_train, y_train)
#
# Score the model
#
print('Best Score:', rs.best_score_, '\nBest Params:', rs.best_params_)
print('Test Accuracy: %0.3f' % rs.score(X_test, y_test))
Resample method for Undersampling or Downsampling Majority Class
In this section, you will learn aboout how to use resample method to undersample or downsample the majority class. Downsampling or undersampling, in the context of handling imbalanced datasets in machine learning, refers to the process of reducing the number of instances in the majority class to balance the class distribution.
Here is the code for undersampling the majority class. In the code below, the majority class (labeled as 1) is downsampled to match the size of the minority class (labeled as 0), using the parameter n_samples=X_imbalanced[y_imbalanced == 0].shape[0]. This ensures the downsampled majority class has the same number of samples as the minority class.
# Downsample the majority class (class 1) to match the minority class count
X_majority_downsampled, y_majority_downsampled = resample(
X_imbalanced[y_imbalanced == 1], # Select only the majority class instances
y_imbalanced[y_imbalanced == 1], # Corresponding labels
replace=False, # No replacement, as this is downsampling
n_samples=minority_class_count, # The number of samples to match the minority class
random_state=123 # For reproducibility
)
# Combine the downsampled majority class with the original minority class
X_undersampled = np.vstack((X_imbalanced[y_imbalanced == 0], X_majority_downsampled))
y_undersampled = np.hstack((y_imbalanced[y_imbalanced == 0], y_majority_downsampled))
Once that is done, the new balanced training / test data set is created and then training and test split get created using the following code.
#
# Create training and test data split
#
X_train, X_test, y_train, y_test = train_test_split(X_balanced, y_balanced, test_size=0.3,
random_state=1, stratify=y_balanced)
The above can be following by usual code for training and scoring the model.
What does Sklearn Resample do?
The resample function in Scikit-learn, found under sklearn.utils, is a utility for randomly sampling data arrays. It is commonly used for modifying the distribution of classes in a dataset, particularly in the context of addressing class imbalances in machine learning. Here’s a breakdown of its main functionalities:
- Random Sampling: resample allows for random sampling of data from arrays, lists, or dataframes.
- Oversampling / upsampling and Undersampling / downsampling: It can be used for both oversampling (increasing the size of a class by sampling with replacement) and downsampling (decreasing the size of a class by sampling without replacement). This is particularly useful in creating balanced datasets from imbalanced ones.
- Replacement Parameter: The replace parameter determines whether the sampling is done with replacement (useful for oversampling) or without replacement (useful for downsampling).
- Sample Size Control: You can control the number of samples to be drawn using the n_samples parameter. This allows for precise control over the size of the sampled dataset.
Conclusions
Here is what you learned about using Sklearn.utils resample method for creating balanced data set from imbalanced dataset.
- Sklearn.utils resample method can be used to tackle class imbalance in the imbalanced dataset.
- Sklearn.utils resample can be used to do both – Under sample the majority class records and oversample minority class records appropriately.
- The Watermelon Effect: When Green Metrics Lie - January 25, 2026
- Coefficient of Variation in Regression Modelling: Example - November 9, 2025
- Chunking Strategies for RAG with Examples - November 2, 2025

Upsampling should only occur on the training set, otherwise resampled training data may also appear in the test dataset.