The drug discovery process is tedious, time-consuming, and expensive. A drug company has to identify the compounds that are most likely to be successful in drug development. The drug discovery process can take up to 15 years with an average cost of $1 billion for each drug candidate that passes clinical trials. With AI and deep learning models becoming more popular in recent years, scientists have been looking at ways to use these tools in the drug discovery process. This article will explore how deep learning generative models (GANs) could be used as a starting point for data scientists to get started drug discovery AI projects!
What is the drug discovery process?
Drug discovery is the process of identifying compounds that could be useful drug candidates. The following are different stages of the drug discovery process:
- Drug identification & design: Drug discovery starts with drug identification. This involves searching for compounds that are active against a biological target of interest, such as an enzyme or other protein involved in disease. Drug design is the next stage where lead compounds are optimized to increase potency and drug-like properties. The drug discovery process can take up to 15 years with an average cost of $1000 million for each drug candidate that passes clinical trials.
- Preclinical research & development (R&D): In the preclinical drug development stage, drug candidates are tested in drug design models. These drug candidates need to be evaluated for efficacy and safety before being considered as drug leads. Before a drug candidate can be evaluated in clinical trials, it has to go through a series of preclinical studies that demonstrate the drug’s safety and efficacy before being tested on humans. This is where drug candidates undergo laboratory and animal testing to answer basic questions about safety.
- Clinical research/trials: Clinical trials involve testing drug candidates in humans for safety and efficacy through phase I to III trials. Phase I involves testing drug candidates in healthy volunteers. Phase II drug development involves clinical trials in patients with the disease that the drug targets. Phase III trials involve large-scale multi-center studies to evaluate the safety and efficacy of drugs approved by the FDA as prescription drug products. In addition, drugs also undergo post-marketing surveillance which involves following drug safety and efficacy after a drug is approved for market.
- Drug review & approval: After clinical trials are over, Drug candidates undergo the drug review process. Drug candidates that show positive results in phase III drug trials need to document their effectiveness and safety, which is reviewed by the government agency before drug approval.
- Drug commercialization: Once drug candidates are approved, drug companies have to complete the drug commercialization process. This involves drug registration, production, and marketing of the drug product before it can be sold in the market for patient use.
What are deep generative models?
Deep generative models are models that are capable of generating synthetic data similar to real observations. Generative adversarial networks (GANs) is one of the most popular deep generative models. GANs are neural networks that consist of two modules, namely; generator and discriminator. The role of each module can be summarized as follows:
- The generator tries to generate synthetic data samples that are similar to real observations in order to fool the discriminator into thinking GANs are deep generative models that have the ability to generate new synthetic examples given existing data points.
- On the other hand, the discriminator tries to differentiate between synthetic and real samples in order to distinguish deep generative models from the non-deep generative models.
Other types of deep generative models include some of the following:
- Gaussian mixture models (GMM): GMM models are used to generate a set of data points from a mixture distribution. In drug discovery, this model can be used to generate molecules that have the same physicochemical properties as existing drug candidates.
- Latent Dirichlet allocation (LDA) models: The Latent Dirichlet Allocation is a generative statistical model that allows sets of observations to be explained by unobserved groups that explain why some parts of the data are similar.
- Hidden Markov models (HMM): Hidden Markov model (HMM) is defined as a set of states that transitions from one state to another. Transitions can be probabilistic or non-probabilistic depending on the model type and drug discovery use cases.
- Variational autoencoders (VAE): Variational autoencoders is a deep generative model that is made up of two parts: an encoder, which compresses real data to latent space; and a decoder, which tries to reconstruct the original input given by encoding.
- Autoregressive models: Autoregressive models are defined as a time series model that consists of a linear combination of previous observations and innovations.
How can deep generative models be used for drug discovery?
With deep learning models becoming more popular in recent years, scientists have been looking at drug discovery from a different perspective. In drug discovery and design, deep learning models can also be used to improve drug candidates with lower toxicity and side effects on patients.
Deep learning generative models (GANs) could be used as a starting point for drug discovery projects. GANs can generate samples of drug-like molecules and provide insights into their properties. This could reduce the overall cost, time and labor involved in drug development process. The following represents different ways in which deep generative models can be used during drug development stages.
- GANs can be used for drug discovery purposes by generating new drug candidates and testing them against a known drug dataset (real data points). GANs models can be used to generate molecules with similar chemical properties as existing drug candidates. GANs can also be used to generate drug candidate molecules with completely different chemical properties. Furthermore, GANs could also be used to identify drug candidates that are similar in chemical properties but have different toxicity and side effects profiles which is important for drug safety assessment purposes. Thus, drug discovery experts can leverage insights into drug candidate molecules’ toxicological profile, their possible adverse effects on the human body for faster drug discovery and design. GANs can be used to predict drug toxicity with the help of autoencoder models that are deep generative models. GAN models could be used to design drug candidates with lower toxicity and better drug safety profiles.
- Quantum GAN: Quantum GAN is a new family of GAN models that can be used for drug discovery purposes. Quantum GAN is based on the principles of quantum mechanics and it tries to minimize the error between drug-like molecules’ ground state (lowest energy level) and deep generative model output. The paper Quantum Generative Models for Small Molecule Drug Discovery was first submitted to ArXiv in January 2021 by June Li, Rasit Topaloglu, Swaroop Ghosh. This model proposes a qubit-efficient Quantum GAN with Hybrid Generator(QGAN-HG). QGAN-HG provides better results than classical GAN because it learns a massive number of molecules by exploring 106 huge chemical spaces with few qubits. QGAN-HG outperforms classic GAN.
- Restricted Boltzmann machines (RBMs): RBM is an unsupervised deep generative model that uses a bipartite approach to learn relations between samples attributes and drug-likeness features. In other words, RBM models are used for drug discovery by training it on a dataset of drug-like molecules and their attributes to learn relations between drug properties.
- Hierarchical Deep Models: Hierarchical deep models are used to generate molecules by using the chemical structure of existing drug leads as a seed molecule and expanding it iteratively while maintaining drug-like properties.
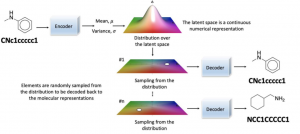
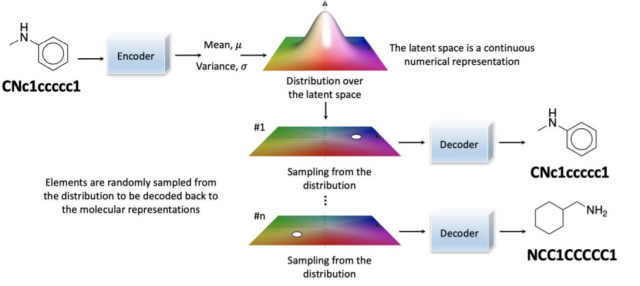
- Generative chemistry with variational autoencoders (VAE): Molecules are encoded into continuous numerical representation in the latent space. Post that, elements are randomly sampled from the distribution and decoded to different numerical representations as shown in the diagram below.
- Generative chemistry with adversarial autoencoders (AAE): AAE is an extension of VAE. It imitates drug molecules’ synthesis process. It is similar to GANs because both AAE and GAN are deep generative models that learn drug-like attributes from datasets of drug leads or drug candidates. AAE enables drug synthesis by generating drug candidate molecules from a drug-like molecule as the seed.
- ACME: ACME is a deep generative model that can be used for drug discovery by learning drug properties from datasets of real drug molecules and their attributes. The paper Generative Adversarial Networks Applied to Organic Chemistry was submitted in June 2020 by M.A. Marriott, J. Amaral-Korytarova, G.M. Golding and Tullio Pozzan.
- Convolutional GAN: ConvGan is a deep generative model that can be used for drug discovery purposes such as generating molecular structures and predicting their properties. The paper CheminGAN: Generating Molecular Structures with Recurrent Neural Networks was submitted in July 2019 by D. Gensert, A.J McCarthy and B.T Winstone.
- Generative chemistry with recurrent neural networks (RNN): RNN is a deep learning model used in drug discovery by learning drug-likeness features from drug datasets. By using drug-likeness features, the model is able to learn several drug-like properties at once. One of RNN’s drug discovery applications takes advantage of its ability to synthesize molecules by generating new ones based on what it learned. This type of deep learning chemoinformatics model is called a generative model where they generate new images (e.g., drug molecules) based on features they learn from data (chemical datasets).
Drug discovery is a complicated process that requires the consideration of many drug-like features. Deep generative models are one tool to help with drug discovery because it can learn drug properties from datasets of drug molecules and their attributes, which gives you more information about your customers’ interests. if you need assistance in implementing these tips, let us know! Do let us know as to what have been some ways that you’ve applied deep learning principles for your drug discovery projects.
- Questions to Ask When Thinking Like a Product Leader - July 3, 2025
- Three Approaches to Creating AI Agents: Code Examples - June 27, 2025
- What is Embodied AI? Explained with Examples - May 11, 2025
I found it very helpful. However the differences are not too understandable for me